scRNA-seq Analysis
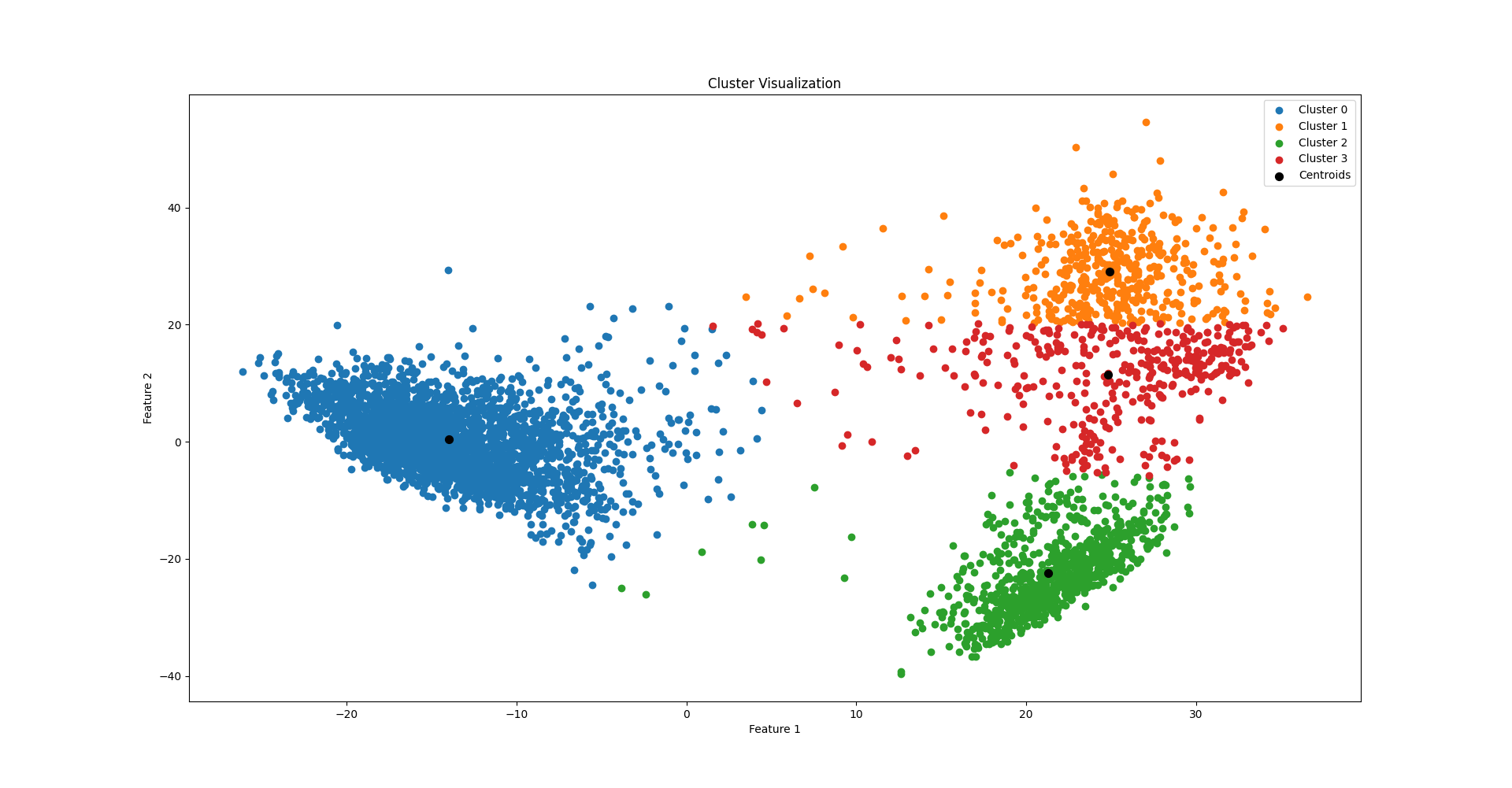
The KMeans clustering algorithm was implemented and applied to a dataset of human pancreas tissue, first reducing its dimensionality using PCA. Utilizing both random and KMeans++ initialization, we explore different numbers of clusters (k ranging from 2 to 10) to identify the optimal clustering configuration, assessed through silhouette coefficients. Lastly, we visualize the clusters in two dimensions using scatter plots to aid interpretation of the clustering outcomes.